 |
FY2021-2025 Grant-in-Aid for Transformative Research Areas (A) “Bottom-up creation of cell-free molecular systems: surpassing nature” |
Research Group
Group A01
The bottom-up creation of artificial cell sensors that enable ultra-sensitive detection of biomarkers and viruses
| Principal Investigator | Shinya Tsukiji (Nagoya Institute of Technology) |
|---|---|
| Collaborator | Kenta Terai (Kyoto University) |

Shinya Tsukiji

Kenta Terai
Research outline
In A01 group, we aim to create artificial cell sensors capable of detecting disease-related biomarkers and viruses with an ultra-high sensitivity. To this end, we will combine various types of molecular materials, including synthetic compounds, lipids, proteins, nucleic acids, and fluorescent probes, and construct liposome-based artificial cell systems that can (i) detect a target analyte biomarker/virus, (ii) transmit the information to the interior of the liposome, and (iii) convert the information to highly amplified (and localized) fluorescence/luminescence signals. Using the systems, we will construct an artificial cell sensor array devise for quantitative digital bioassay. We will also apply the artificial cell sensors as novel diagnostic tools for detecting biomarkers/viruses ex vivo and in vivo. Specifically, we will focus on the following three projects.
- Bottom-up construction of ultra-sensitive artificial cell sensors for proteolytic and hydrolytic enzyme biomarkers
- Bottom-up construction of ultra-sensitive artificial cell sensors for non-enzymatic biomarkers and viruses
- Application of artificial cell sensors as new diagnostic tools
During these processes, we will establish methodologies and chemical principles to design artificial cell-like systems with spatially-controlled, multistep, signal transduction/amplification functions. We will conduct this project in collaboration with B01, B02, and E01 groups.
Keywords
artificial cell, synthetic functional molecules, signal amplification, sensor
References
JACS 2013, 135, 12684., Biochemistry, 2020. 59, 205., ACS Chem. Biol. 2021, 16, 1557.
Group B01
Bottom-up creation of fully reconstituted virus- and bacteria-like particles
| Principal Investigator | Katsutoshi Hori (Full Professor, Nagoya Univ.) |
|---|---|
| Collaborator | Rie Wakabayashi (Assistant Professor, Kyushu Univ.) |
| Collaborator | Ryo Ueno (Specially Appointed Assistant Professor, Okayama Univ.) |

Katsutoshi Hori

Rie Wakabayashi

Ryo Urano
Research Outline
Both viruses and bacterial cells are elaborate molecular systems created by nature, and Group B01 is going to develop a liposome-based artificial particle creation technology to construct practical bacteria-like particles BLP (≃1 µm) and virus-like particles VLP (≃0.1 µm) from biomolecules and artificial materials with the assistance of computational science.
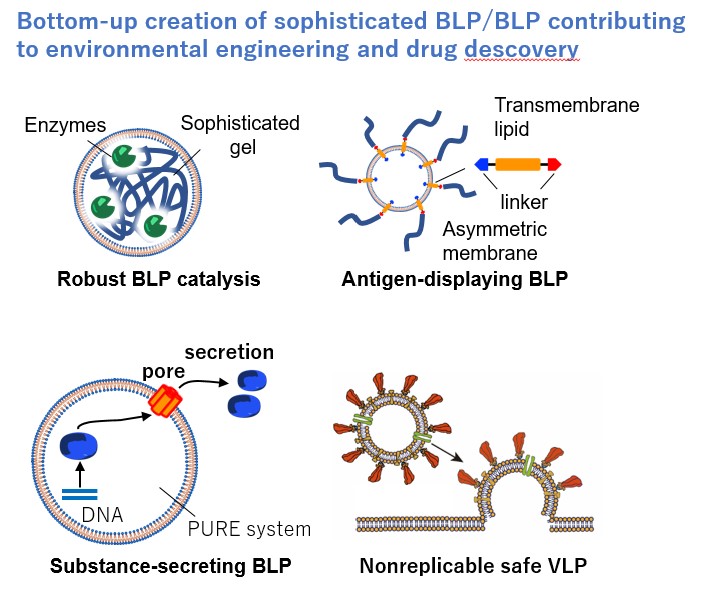
We will create practical molecular systems with a view to the exit of “investigation” and “protection”, such as immobilized BLP catalysts with excellent desiccation resistance and physical strength for air purification, membrane-fusion-type VLPs and substance-secreting BLPs for drug delivery systems, and safe VLPs/BLPs for elucidation of infection mechanisms and drug discovery. We will establish the methodology for constructing molecular systems by controlling the spatio-temporal arrangement of interacting molecules through a combinatorial search of building factors, such as particle size, composition, molecular orientation, assembly order, and fluidity. By comparing VLP (nano-space) and BLP (micro-space), we will elucidate the formation mechanisms of structural complexity and order across scales and derive the theories for constructing molecular systems.
Keywords
bacteria-like particle、virus-like particle、molecular interaction
References
J.Am. Chem.Soc. 141, 2019, 19058; Green Chem. 22, 2020,1258; J. Colloid Interface Sci. 606, 2022, 628; Microbial. Spectr. 9,2021, e0025
Group B02
Bottom up creation of molecular systems that enable the in vitro evolution of protein complexes
| PI | Tomoaki Matsuura (ELSI, Tokyo Institute of Technology) |
|---|---|
| Collaborator | Natsuko Miura (Osaka Metropolitan University) |
| Collaborator | Daisuke Kiga (Waseda University) |

Matsuura, T.

Miura, N.

Kiga, D.
Research Outline
In B02 group, we will bottom-up construct cell-free molecular systems to realize in vitro evolution that cannot occur in nature. We will use these systems to create molecules that will contribute to social implementation. Specifically, we will tackle the following three issues.
- Construct molecular systems that enables in vitro evolution of membrane proteins, and derive membrane protein variants that contribute to drug discovery and chemogenetics.
- Construct molecular systems that enables in vitro evolution of proteins using a cell-free system with an artificial codon table, and generate oxidation-resistant enzymes that can be implemented in biosensors and etc.
- Construct molecular systems that enable in vitro evolution of artificial enzyme assemblies using cell-free systems, and derive highly efficient production systems for industrially important chiral compounds.
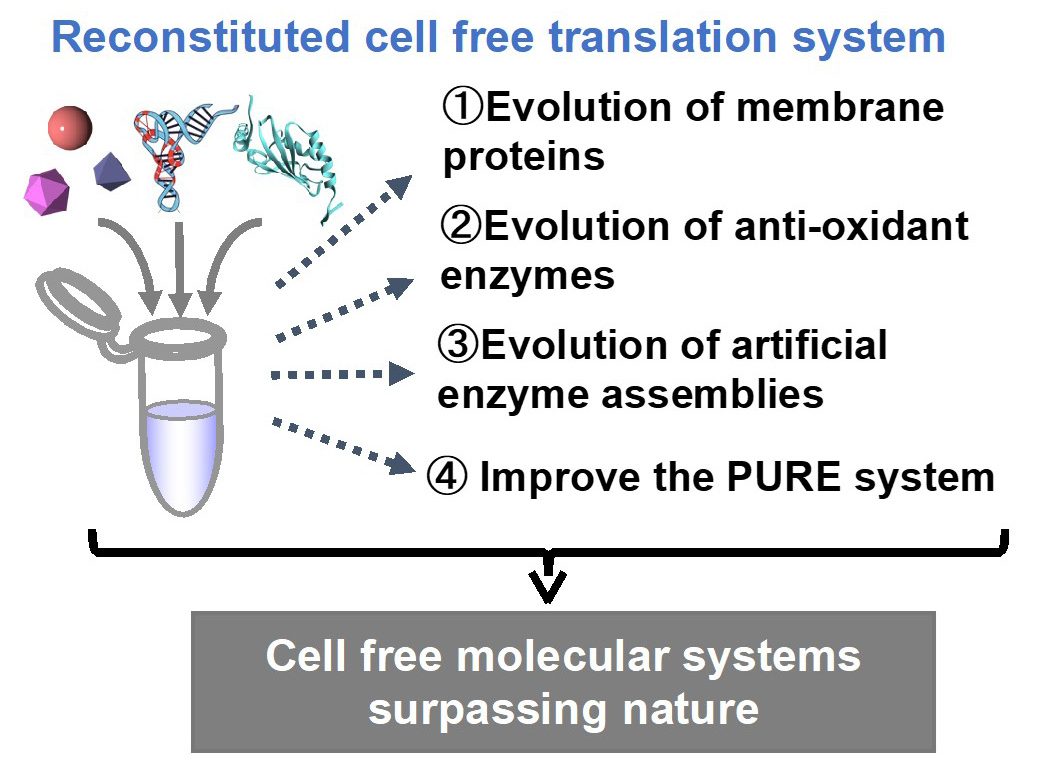
In addition, we will construct new cell-free molecular systems in collaboration with groups A01, B01, and D01. During these processes, we will conduct interaction analyses, and aim to clarify guidelines for the constructing valuable cell-free molecular systems.
Keywords
Directed evolution, cell-free system, interaction analysis
References
Proc Natl Acad Sci US A, 2013, 110, 16796; ACS Synth Biol, 2018, 7, 2537; Eukaryot. Cell, 2013, 12(8), 1106
Group C01
Bottom-up construction of de novo cell-membrane system
| Principal Investigator | Ryuji Kawano(Tokyo University of Agriculture and Technology・Professor) |
|---|---|
| Collaborator | Izuru Kawamura(Yokohama National University・Associate Professor) |

Ryuji Kawano

Izuru Kawamura
Research Outline
In group C01, we systematize artificially designed molecules with de novo to construct an artificial cell-membrane system from the bottom up. In addition to phospholipids and sterols, the membranes of living cells are filled with a large number of membrane proteins. Membrane proteins are important for the exchange of materials, energy, and information between inside and outside the cell.
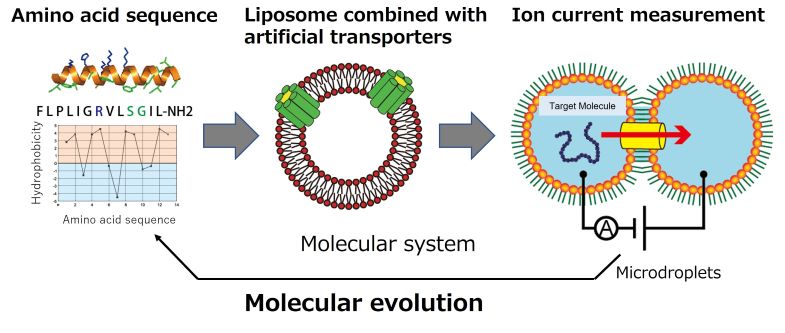
In this project, we will artificially construct a cell membrane with designed nanopore molecules with nano-sized pores, as well as membrane transporter molecules aiming to establish a de novo cell membrane molecular system that systematizes multiple molecules of lipids, sterols, and membrane transporters. In order to achieve this goal, this project group plans to construct micro-sized artificial cell-membrane systems based on microfabrication, microfluidics, electrochemical measurement technology, and solid-state NMR spectroscopy. In addition, we will also construct new artificial cell membrane molecular systems in collaboration with groups A01, B01, B02, and D01.
Keywords
Nanopore、Lipid bilayer、Microfluidics、Solid-state NMR
References
Nature Nanotech. 2021; Micromachines 2020 (Review); ChemPhysChem 2018 (Review); Nature Chem 2016
Group D01
Optimization of cell-free molecular systems with computational science approaches
| PI | ABURATANI Sachiyo (CBBD-Open Innovation Lab, AIST) |
|---|---|
| Co-PI | SHIRAI Tomokazu (Cell Factory Research Team, RIKEN) |
| Co-PI | HONDA Kohsuke (International Center for Biotechnology, Osaka University) |

PI: ABURATANI Sachiyo

Co-PI: SHIRAI Tomokazu

Co-PI: HONDA Kohsuke
Research outline
Our group aims to develop platform technologies available for the optimization of various cell-free molecular systems by integrating biochemical approaches and computational science. Our research scheme consists of (i) rational selection of building blocks of a cell-free molecular system from database, (ii) computational modeling, design, and optimization of the system, and (iii) actual construction and implementation of the system. As proof-of-concept studies, we develop and optimize following cell-free molecular systems.
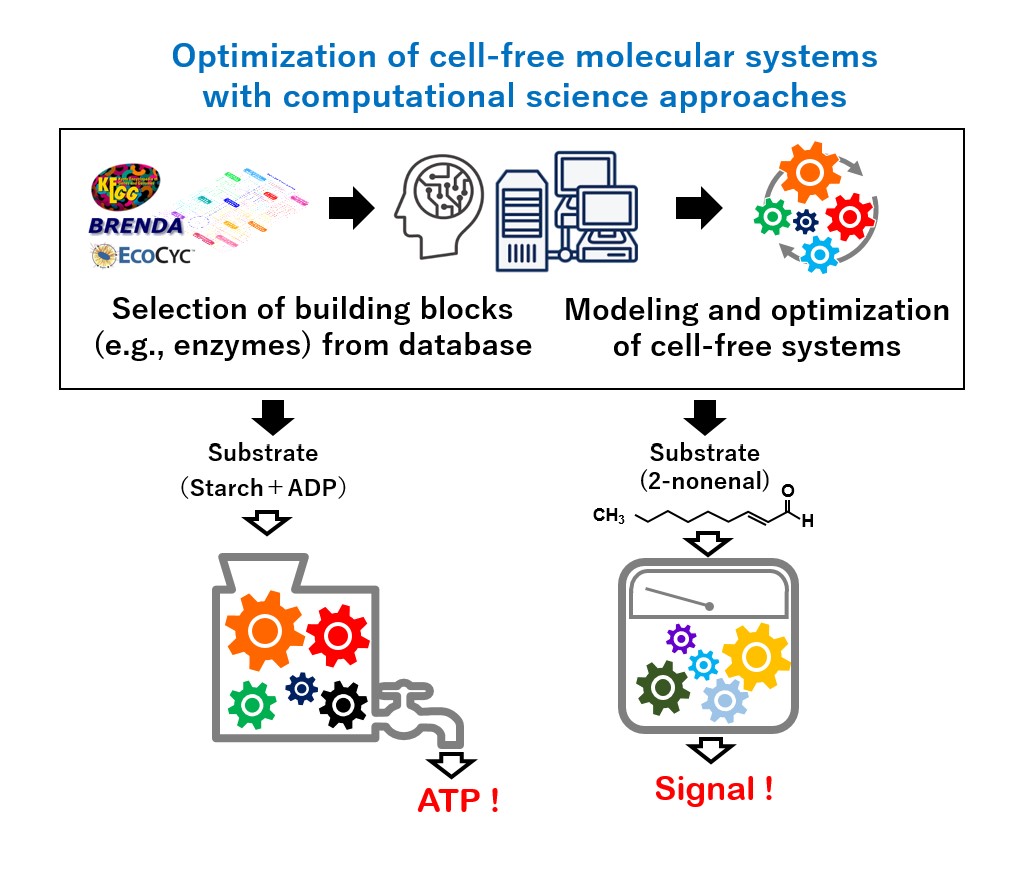
- Enzymatic determination system of body-odor compound
- ATP regeneration/production system with artificially designed glycolysis
- Rate-enhancement technology for multi-step enzyme reaction using self-assembly peptide
Besides these systems, we apply our optimization technologies to more variety of cell-free molecular systems, including those developed by other groups in this research area.
Keywords
System optimization, Causal inference in statistics, 2-nonenal, ATP
Related papers
J Phys Cont Ser., 1391, 012047; Microb. Cell Fact., 18(1):1 24; Appl. Microbial. Biotechnol. 103
Group E01
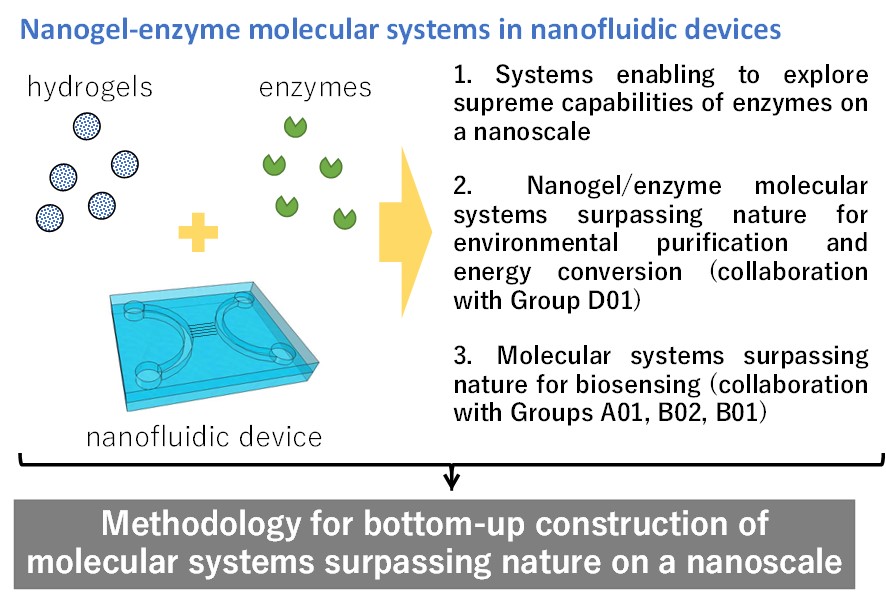
Bottom-up construction of nanogel-enzyme molecular systems using nanofluidic devices
| Principal Investigator | Yan XU, Associate Professor, Osaka Prefecture University |
|---|---|
| Co-Investigator | Madoka TAKAI, Professor, The University of Tokyo |

Yan XU

Madoka TAKAI
Research Outline
In this study, we aim to develop methodology for nanoscale bottom-up construction of molecular systems surpassing nature by using components such as polymer hydrogels, enzymes, nanofluidic devices. We attempt to elucidate multi-parameter interactions among the enzymes and the polymer hydrogels confined in the nanofluidic devices. Meanwhile, we attempt to explore a nanofluidic space enabling to maximize the enzyme activity, with the help of computational science developed by Group D01. The new methodology will allow for the bottom-up construction of nanogel-enzyme molecular systems with surpassing nature capabilities for applications in the fields of environmental purification, energy conversion, and biosensing, in collaboration with Groups A01, B02, and B01. The realization of such molecular systems would finally contribute to the protection of our human beings and the earth.
Keywords
nanofluidic devices, polymer hydrogels, enzymes, interaction analysis
References
Adv. Mater., 2016, 28, 2209; Adv. Mater., 2018, 30, 1702419; ACS Appl. Polym. Mater. 2021, 3, 631.

